Daniel Vella and Jean-Paul Ebejer (2023)
Highlighted by Jan Jensen
This paper is an update and expansion to this seminal paper by Pande and co-workers (you should definitely read both). It compares the ability to distinguish active and inactive compounds for few-shots methods to more conventional approaches for very small datasets. It concludes that the former outperform the latter for some data sets and not for others, which is surprising given that few-shot methods are designed with very small data sets in mind.
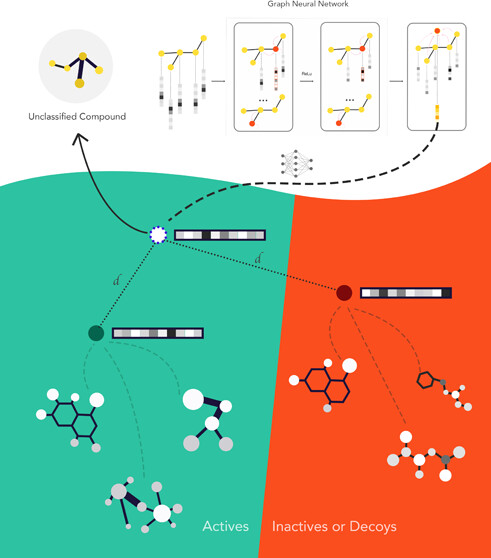
Few shot methods learn a graph-based embedding that minimizes the distance between samples and their respective class prototypes while maximizing the distance between samples and other class prototypes (where prototypes often are the geometric center of a group of molecules). The training set, which is composed of a "query set" that you are trying to match to a "support" set support set is typically small and changes for each epoch (which is now called episodes) to avoid overfitting.
In this paper, the largest support set was composed of 20 molecules (10 actives and 10 inactives) sampled (together with the query set) from a set of 128 molecules with a 50/50 split of actives and inactives. The performance was then compared to RF and GNN models trained on 20 molecules.
My main takeaway from the paper was actually how well the conventional models performed. Especially given the fact that the conventional models actually had smaller training set, since the few-shot methods saw all 128 molecules during training over the course of the training, whereas the conventional methods only saw a subset.

This work is licensed under a Creative Commons Attribution 4.0 International License.