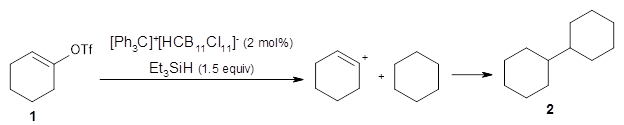Jiaxuan You, Bowen Liu, Rex Ying, Vijay Pande, Jure Leskovec (2018)
Highlighted by Jan Jensen

Ever since Alán Aspuru-Guzik and co-workers published their seminal paper there has been a flurry of activity on generative models, which is not surprising given that they offer a radically new alternative to screening chemical libraries as a way to discover new molecules.
Almost all the new efforts on generative models have been based on adapting machine learning techniques used for natural language processing to text-based representation of molecules, i.e. SMILES strings. While very promising the SMILES syntax has some quirks which makes them hard to predicts efficiently. One solution is to change the syntax to be more ML-friendly, but this has yet to be tested for generative models.
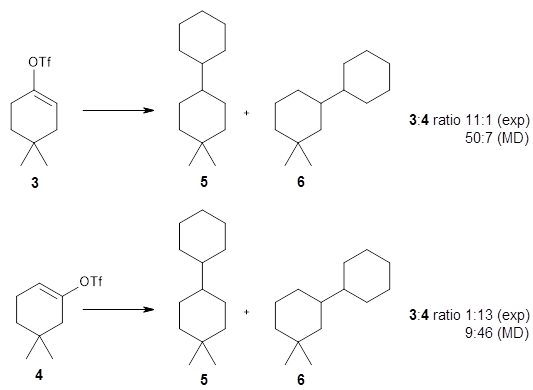
Another option is to work with a graph (i.e. atoms and bonds) representation of the molecule and this paper is the first I've seen that does that for an ML-based generative model. In this case the ML method is reinforcement learning where the addition of each atom is treated as an action which can be trained to towards a particular outcome, here molecules with certain properties. This approach seems to outperform the SMILES based approaches for the prediction of some properties.
The code is available here.

This work is licensed under a Creative Commons Attribution 4.0 International License.

Ever since Alán Aspuru-Guzik and co-workers published their seminal paper there has been a flurry of activity on generative models, which is not surprising given that they offer a radically new alternative to screening chemical libraries as a way to discover new molecules.
Almost all the new efforts on generative models have been based on adapting machine learning techniques used for natural language processing to text-based representation of molecules, i.e. SMILES strings. While very promising the SMILES syntax has some quirks which makes them hard to predicts efficiently. One solution is to change the syntax to be more ML-friendly, but this has yet to be tested for generative models.
Another option is to work with a graph (i.e. atoms and bonds) representation of the molecule and this paper is the first I've seen that does that for an ML-based generative model. In this case the ML method is reinforcement learning where the addition of each atom is treated as an action which can be trained to towards a particular outcome, here molecules with certain properties. This approach seems to outperform the SMILES based approaches for the prediction of some properties.
The code is available here.

This work is licensed under a Creative Commons Attribution 4.0 International License.