Markopoulos, G.; Grunenberg, J. Angew. Chem. Int. Ed. 2013, 52, 10648
Contributed by Steven Bachrach.
Reposted from Computational Organic Chemistry with permission
Contributed by Steven Bachrach.
Reposted from Computational Organic Chemistry with permission
Can one identify a labile bond in a molecule without computing activation barriers? Markopoulos and Grunenberg suggest that examination of the bond length and its associated relaxed force constant might provide some guidance.1
The relaxed force constant comes from identifying the force constant for some coordinate while allowing for other coordinates to relax. Badger’s rule relates the (normal) force constant to bond distance (k = a/(req – d)3). For a series of 36 molecules, covering 71 C-C single bonds, Badger’s rule fits the data well, except for a set of molecules which undergo rapid Cope rearrangements (like bullvalene and semibullvalene). For these molecules, the relaxed force constants are much lower than Badger’s rule predicts, and indicates a weakened bond. This gives rise to their low activation barriers.
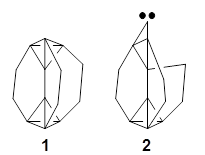
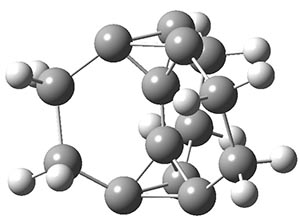
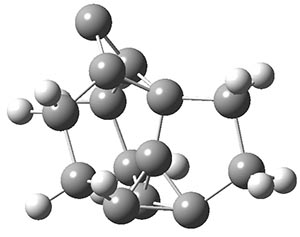
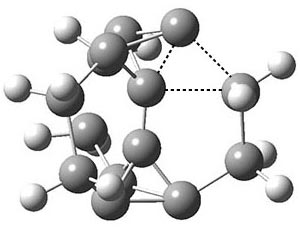
Another example is provided with the highly strained polycyclic hydrocarbon 1. This compound is predicted (B3LYP/6-31G(d)) to undergo a [1,2]-shift to give the carbene 2 (see Figure 1), and this is extremely exothermic: -105.7 kcal mol-1, reflecting the enormous strain of 1. The barrier, through TS1 (Figure 1), is only 6.7 kcal mol-1. This rearrangement was predicted by examining the relaxed force constants which identified a very weak bond, despite a short bond distance of 1.404 Å. It is unlikely that without this guidance, one would have predicted that this short bond is likely to rupture and produce this particular product.

1
|
2
|
TS1
| |
Figure 1. B3LYP/6-31G(d) optimized structures of 1, 2, and TS1.
References
(1) Markopoulos, G.; Grunenberg, J. "Predicting Kinetically Unstable C-C Bonds from the Ground-State Properties of a Molecule," Angew. Chem. Int. Ed. 2013, 52, 10648-10651, DOI: 10.1002/anie.20130382.
InChIs
1: InChI=1S/C14H12/c1-2-8-11-5-3-9-7(1)10(9)4-6-12(8,11)14(8,11)13(7,9)10/h1-6H2
InChIKey=LNBZAENQMFDBJW-UHFFFAOYSA-N
InChIKey=LNBZAENQMFDBJW-UHFFFAOYSA-N
2: InChI=1S/C14H12/c1-3-11-12-4-2-9-7-8(1,9)10(9)5-6-13(11,12)14(10,11)12/h1-6H2
InChIKey=UKVODHRLGFPZPT-UHFFFAOYSA-N
InChIKey=UKVODHRLGFPZPT-UHFFFAOYSA-N

No comments:
Post a Comment